Introduction
Intravascular optical coherence tomography (IVOCT) enables detailed in vivo visualisation of atherosclerotic plaque with high resolution and accurate characterisation of different types of tissue composition1. This information is instrumental for plaque-specific lesion preparation and for appropriate selection of the therapeutic strategy during IVOCT-guided intervention2,3. However, plaque characterisation is currently challenging, time-consuming, difficult to systematise, based on subjective interpretation of the operators and largely dependent on their expertise, thus posing problems for reproducibility.
Artificial intelligence (AI) might be useful to standardise and automate plaque characterisation in interpreting IVOCT images. Deep learning (DL) is a data-driven AI algorithm to extract regular patterns from the observation of data by training a designed model. After training, the model learns features that can be used to make predictions in unexplored data sets. Convolutional neural network (CNN) is a specific architecture in DL that has excellent performance in image processing. Several studies have attempted automatic characterisation of atherosclerotic plaques in coronary arteries using CNN, following different approaches4,5,6.
The aim of this study was to develop a deep convolutional network for comprehensive plaque characterisation, trained on a large diverse data set of IVOCT images comprising varied anatomic morphologies and clinical scenarios, to provide both qualitative characterisation and quantification of plaque components for clinical use.
Methods
STUDY DESIGN AND STUDY POPULATION
This was a retrospective, post hoc, multicentre, international study to appraise the accuracy and reproducibility of a novel diagnostic method for automatic plaque characterisation in IVOCT.
For the development of the CNN model, patients and lesions from three different IVOCT studies7,8,9 were included in the current post hoc analysis. Briefly, the studies included patients from five international centres located in Australia, USA, Japan, Spain and China, in both a retrospective7,8 and a prospective fashion9, with IVOCT performed for the evaluation of stable coronary lesions. Exclusion criteria were aorta-ostial lesions, bypass graft lesions, patients with moderate or severe valvular heart disease, acute coronary syndrome <72 hours attributed to the imaged vessel and chronic total occlusion in any other vessel. All IVOCT pullbacks were acquired with frequency-domain OCT systems of C7-XR™ or OPTIS™ (Abbott Vascular, Santa Clara, CA, USA) using a non-occlusive technique10. The institutional review boards of each individual centre approved the protocol of the studies, and all patients provided informed consent for enrolment in the institutional database for potential future investigations.
For the independent external validation, a different data set of 300 IVOCT images, provided by three international core labs, was used. Five patients were studied using the Lunawave® OFDI system (Terumo Corporation, Tokyo, Japan), while the rest were studied using the frequency-domain OCT OPTIS system.
DATA ANNOTATION
Ground truth was generated by labelling nine objects by experienced OCT analysts. The detailed annotation strategy is available in Supplementary Appendix 1.
DEEP CONVOLUTIONAL MODEL ARCHITECTURE: DESIGN AND TRAINING (Supplementary Appendix 2)
A U-shaped encoder–decoder architecture was designed, consisting of a contracting path for high-level feature extraction, an expansion path to produce full resolution segmentation, and vertical and horizontal feature bridges to preserve detailed spatial information (Supplementary Figure 1). The model was fed with pseudo-3D input by stacking consecutive IVOCT cross-sections as separate colour channels to integrate the spatial information. A hybrid loss function of multi-class cross-entropy loss and focal Tversky loss was used to address the problem of class imbalance (Supplementary Appendix 3). The detailed training strategy and ablation experiments to test the rationale of the CNN design are available in Supplementary Appendix 4 and Supplementary Appendix 5.
MODEL DEVELOPMENT AND INTERNAL EVALUATION
The annotated pullbacks were randomly divided into training data set and testing data set, in a proportion of 9 to 1, strictly avoiding repetition of pullbacks in different data sets. The training set was used for the model development, of which 10% of the data set was separated for the hyper-parameter optimisation. After the model was fully developed, the testing data set was used for the internal evaluation of the model performance. The agreement between predictions and ground truth was evaluated by means of the Dice coefficient, calculated per category and then averaged over categories. The purity and completeness of positive predictions relative to the ground truths were reported as precision and recall, respectively.
Plaque burden (PB) was calculated as the area between the estimated internal elastic lamina (IEL) contour and lumen contour, divided by IEL area and multiplied by 100%11. The accuracy in segmentation of the IEL was appraised as the agreement in PB between the model prediction and the ground truth.
EXPERT CONSENSUS AND INDEPENDENT VALIDATION
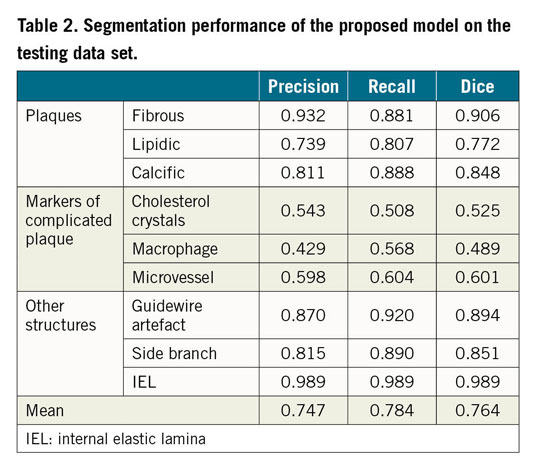
The external validation of the model was performed on a different data set than the one used for the model development or internal evaluation. A total of 300 IVOCT images with different atherosclerotic plaque morphology and composition, acquired during clinical practice, were provided by three international core labs, each one providing 100 images (from 10 patients) with delimited regions (Figure 1). Regions with poor signal penetration or insufficient quality, precluding adequate visualisation and analysis, were flagged with an arch-shaped annotation (Figure 1, region 3). Four experienced OCT readers (A. Maehera, Z. Ali, H. Jia, N. Holm) from the three core labs (CRF, New York, NY, USA; iMcorelab, 2nd Affiliated Hospital of Harbin Medical University, Harbin, China; Aarhus University Hospital, Skejby, Denmark) participated in the evaluation and labelled the regions with the corresponding tissue components, having access to the original images but blinded to each of the other analysts. If the evaluator considered that the delimited region had more than one tissue type, the region was then subdivided into multiple regions. According to the evaluation by the three core labs, each region was classified as “unanimity”, “one core lab disagrees” or “all three core labs disagree”. Supplementary Figure 2 shows representative cases with unanimity and disagreement among core labs. Consensus was defined as agreement between ≥2 core labs, i.e., first two categories of the classification, and the consensus label for the region was accepted as reference for the independent validation.
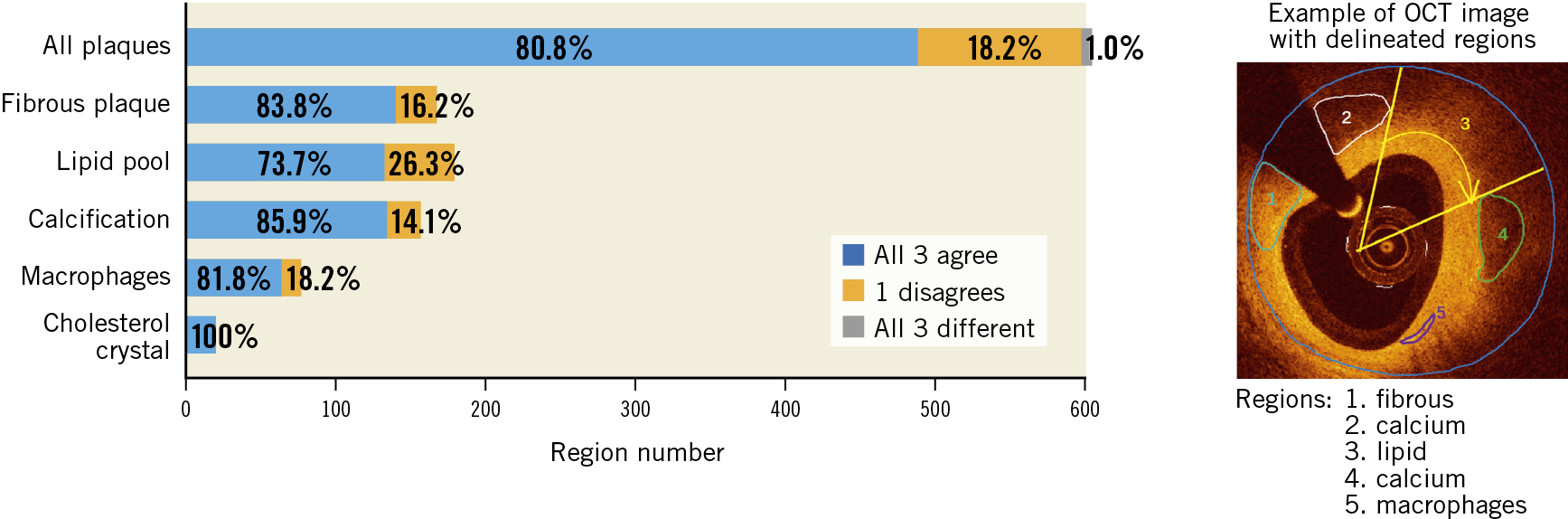
Figure 1. Inter-core lab agreement and variability on plaque characterisation. Example of an IVOCT image with delimited regions (marked by numbers) and results of plaque characterisation, stratified by the different tissue components, as determined by the three core labs. There is good agreement for the majority of calls, since all three or at least two core labs agreed on the diagnosis.
The proposed deep convolutional model was integrated into the OctPlus software (Pulse Medical Imaging Technology, Shanghai, China) for real-time analysis of IVOCT pullbacks (Figure 2), displaying plaques in both 2D cross-sectional and 3D views (Figure 2A). The software quantified plaque area, arc degree and the different tissue proportions. An independent validation of the proposed CNN model within the software was performed, using the consensus regions as reference. Given that some plaque regions were flagged by arch-shaped annotations without complete boundaries, pixel-wise evaluation by Dice coefficient was not applicable for the external validation. Model performance was reported by the means of accuracy. Correct plaque characterisation was defined as a >80% overlapping portion between the prediction of the software and the core lab consensus in area or arc circumference. Sensitivity analysis on the impact of the overlapping portion on the diagnostic accuracy was performed by changing the overlapping portion from 80% to 90%.
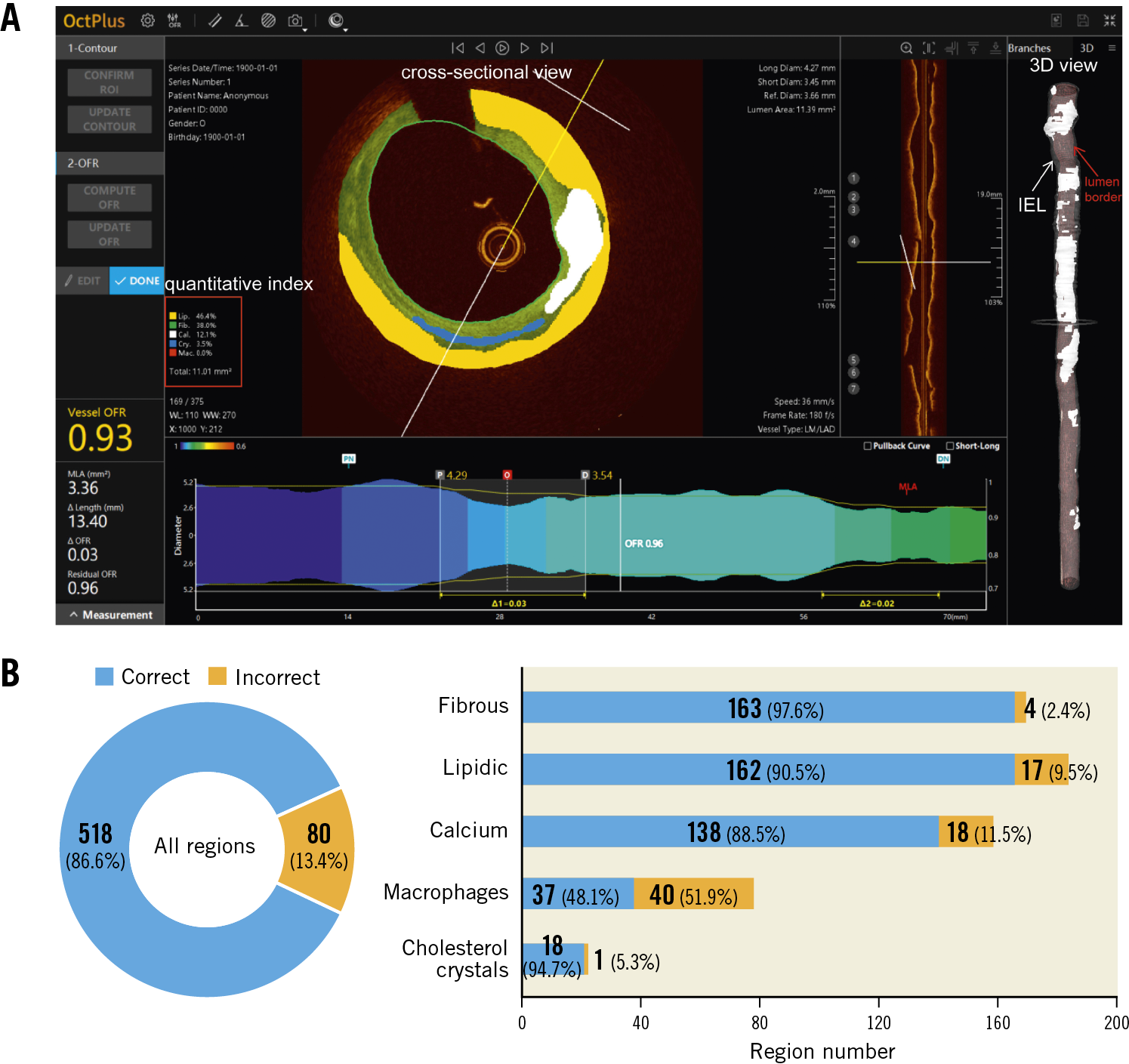
Figure 2. The proposed AI model was integrated into the OctPlus software and externally validated. A) Screen shot of plaque quantitative assessments by the software. 3D mapping of calcifications between IEL (white arrow) and lumen contour (red arrow) is shown on the right side. B) Diagnostic accuracy of the software, stratified by the different plaque components, taking the inter-core lab consensus as standard reference. AI: artificial intelligence; IEL: internal elastic lamina
STATISTICAL ANALYSIS
A normality test was performed using the Shapiro-Wilk test. Continuous variables are presented as mean±SD or median (interquartile range), as appropriate, whereas categorical variables are presented as counts and percentages. Correlation between ground truth and model predictions was evaluated using Pearson or Spearman correlation tests, as appropriate. Agreement between groups for continuous variables was assessed by means of Bland-Altman analysis and intraclass correlation coefficient for the absolute agreement (ICCa), whilst the kappa coefficient was used for categorical variables. A two-sided p-value ≤0.05 was considered statistically significant. A confidence level of 95% (95% CI) was used to estimate the plausible range of values. Statistical analysis was performed using SPSS, Version 23.0 (IBM Corp., Armonk, NY, USA).
Results
STUDY POPULATION CHARACTERISTICS
A total of 509 OCT pullbacks from 391 patients were analysed, resulting in 10,517 and 1,156 cross-sections for the training and testing data sets, respectively (Figure 3). Patient baseline clinical characteristics are presented in Table 1.
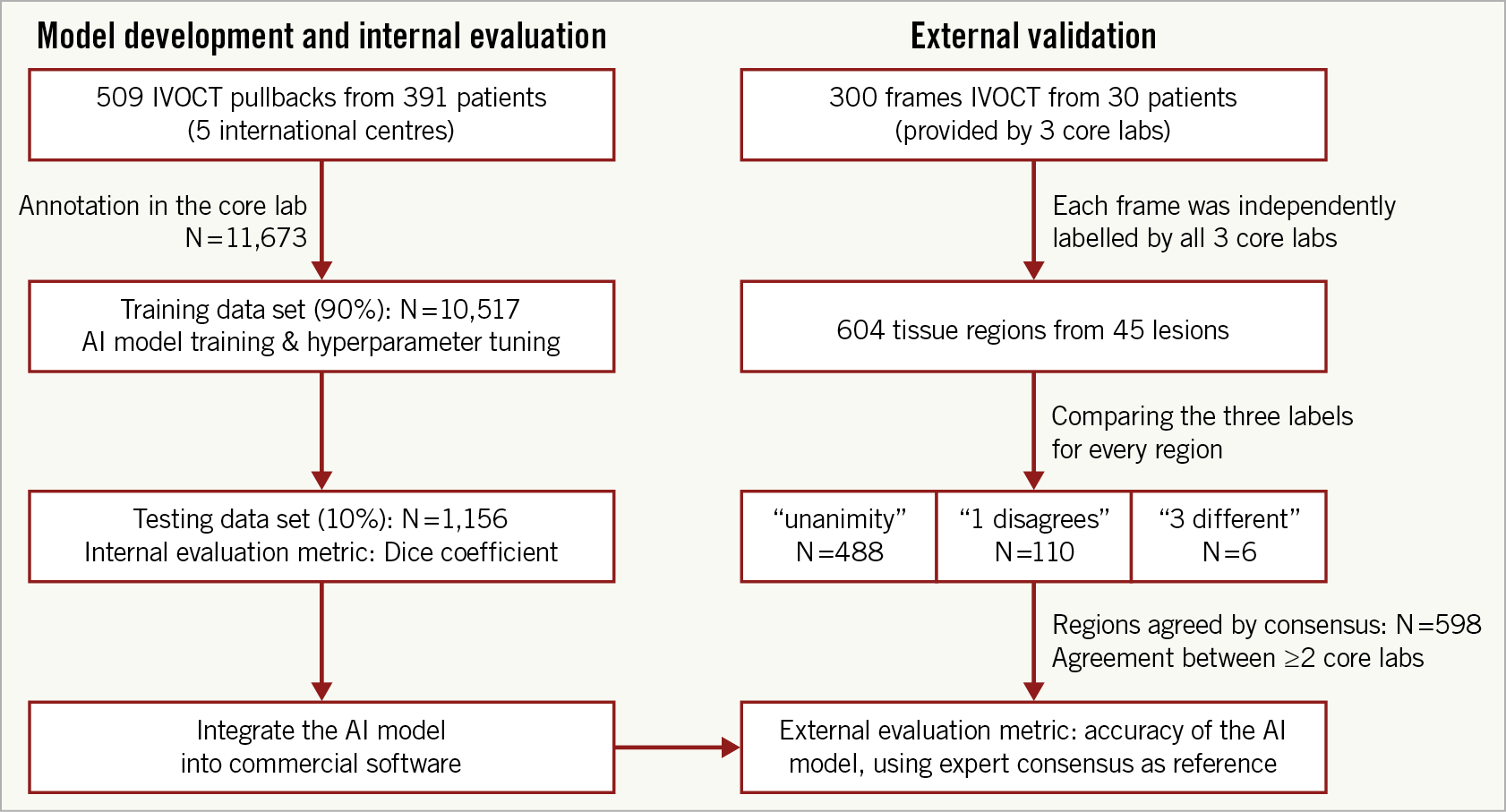
Figure 3. Study flow chart. The utility of the different data sets for internal evaluation (left column) and external validation (right column) is indicated.
INTERNAL EVALUATION
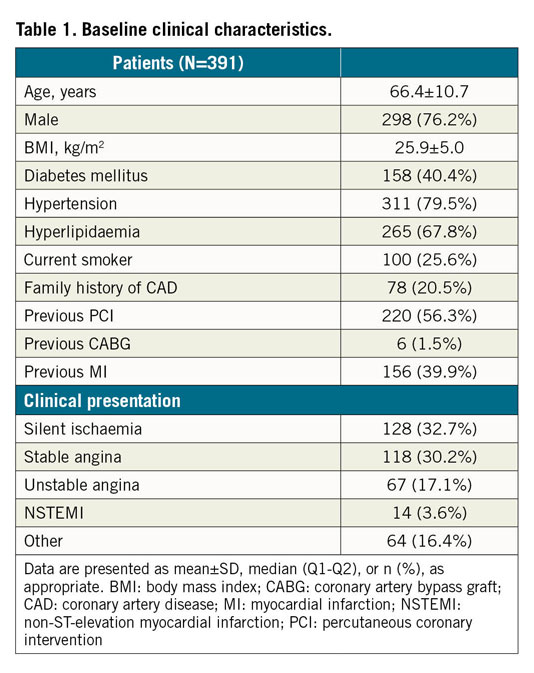
The model performance on the testing data set is summarised in Table 2. The model performed the best segmentation on fibrous plaque (Dice=0.906), followed by calcific (Dice=0.848) and lipidic plaque (Dice=0.772). For the segmentation of markers of inflammation/complicated plaque, the model performed best on microvessels (Dice=0.601), followed by cholesterol crystals (Dice=0.525) and macrophages (Dice=0.489). Segmentation of non-tissue structures (i.e., guidewire and side branches) also achieved high performance (Table 2). Among quantitative parameters, the PB assessed by the model correlated very well with the ground truth (R2=0.98, p<0.001) (Figure 4), with a mean difference of 0.35±2.2% and an ICCa of 0.99 (95% CI: 0.98-0.99).
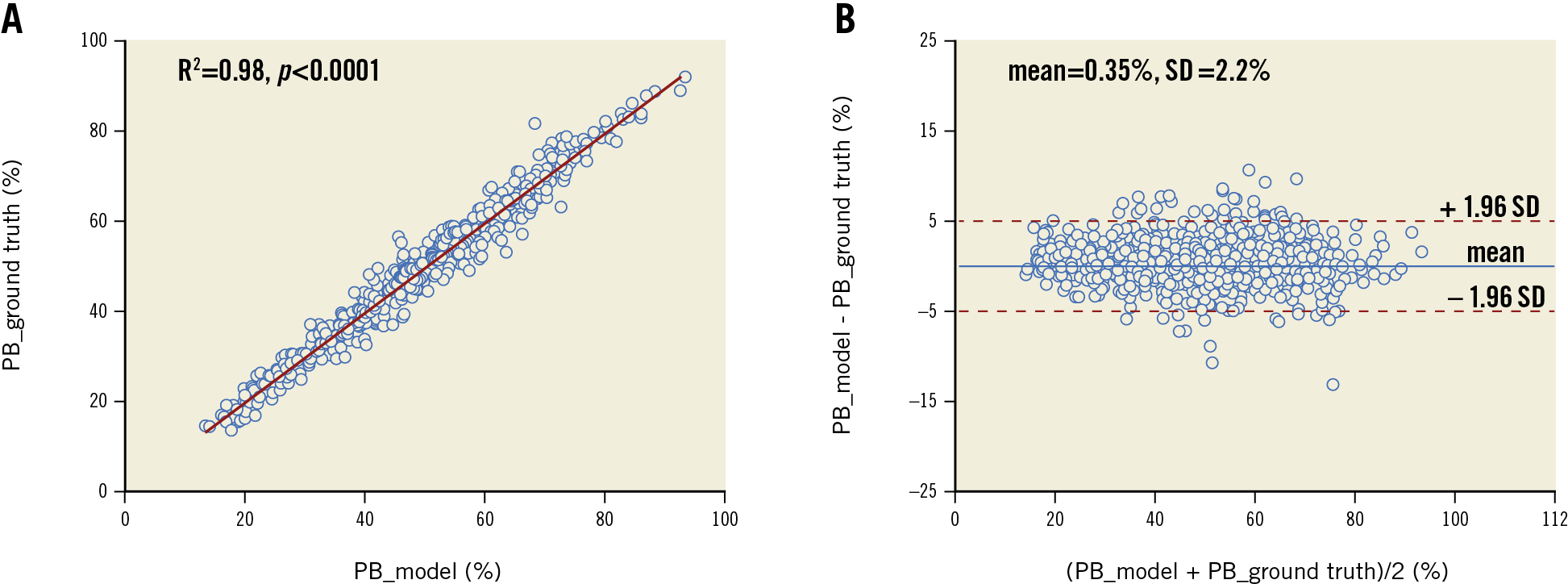
Figure 4. Agreement of plaque burden (PB) between AI predictions and manual annotations. A) Correlation in PB between model prediction and ground truth. B) Bland-Altman analysis. Middle blue line: mean difference; red dotted lines: mean±1.96 SD. PB: plaque burden
Figure 5 shows examples of the model predictions in different challenging scenarios. The model had excellent performance on cases with insufficient blood clearance (row A), complex calcification (row B) or large lipidic burden (row C), while displaying certain ability in segmenting different inflammatory markers, like cholesterol crystals, macrophages or microvessels (row D).
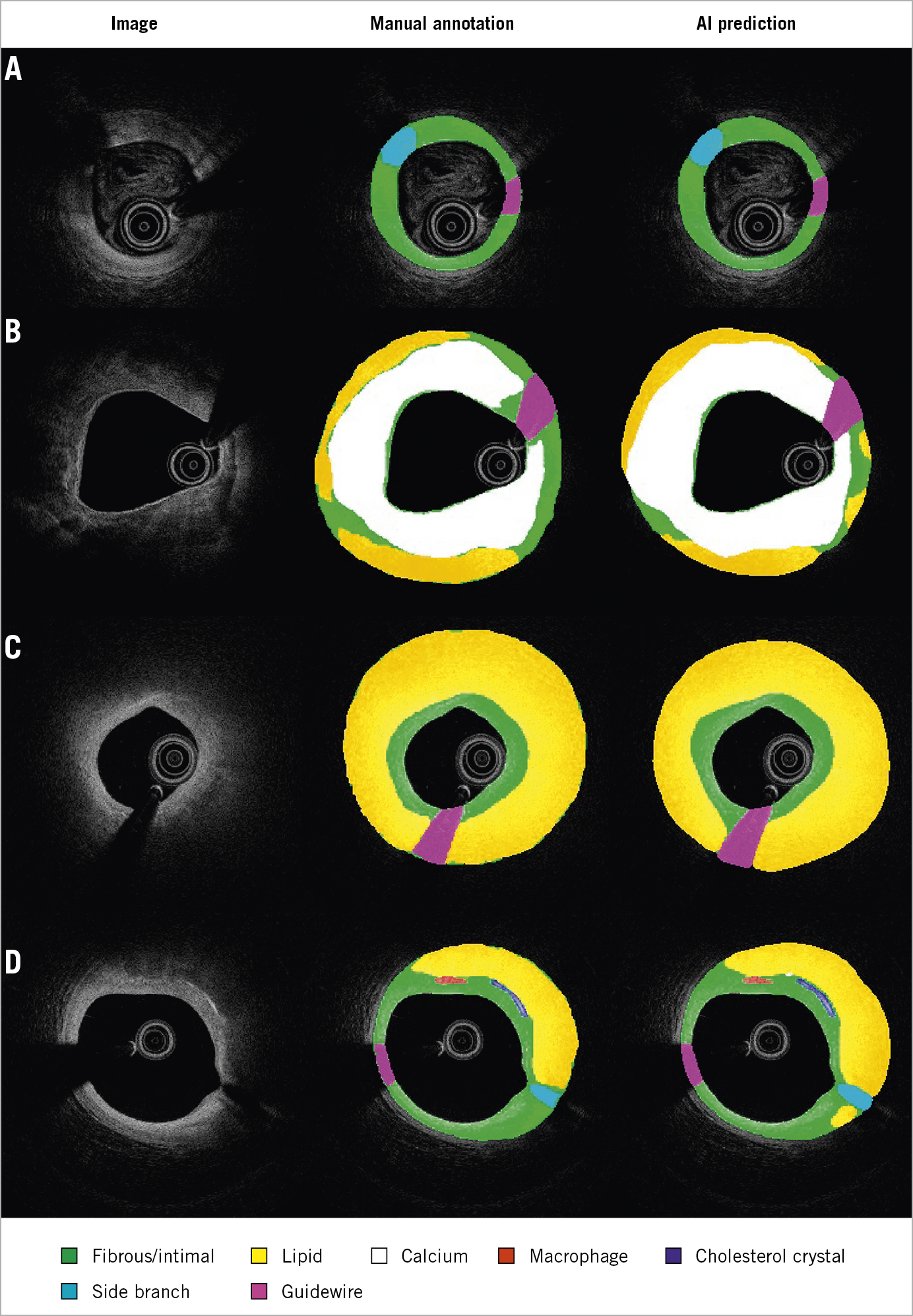
Figure 5. Segmentation in different challenging situations. Images with suboptimal quality (A), heavy calcification (B), plaques with large lipidic pool (more than two quadrants) (C), inflammatory markers (D).
INTER-CORE LAB AGREEMENT AND VARIABILITY IN PLAQUE CHARACTERISATION
For the external validation, a total of 45 lesions, including 10 long lesions (>28 mm) and 6 diffuse lesions from the 30 IVOCT examinations were analysed, resulting in 604 plaque regions labelled by the core labs (Figure 3). The median lesion length and minimal lumen area of the lesions were 20.40 (10.90-33.05) mm and 2.00 (1.36-3.00) mm2, respectively. Consensus on plaque characterisation was reached in 598 (99% [95% CI: 97.8-99.6%]) regions (Figure 1); unanimity among core labs was observed in 488 (81% [95% CI: 77.5-83.7%]) regions, and agreement between two core labs in 110 (18% [95% CI: 15.3-21.5%]) regions. Unanimity among core labs was most frequently observed for cholesterol crystals (100% [95% CI: 82.4-100%]), followed by calcific plaque (85.9% [95% CI: 79.5-90.6%]), fibrous plaque (83.8% [95% CI: 77.5-88.7%]), macrophages (81.8% [95% CI: 71.6-88.9%]), and lipidic plaque (73.7% [95% CI: 66.8-79.7%]). The data on agreement between individual core labs with consensus are available in Supplementary Appendix 6 and Supplementary Figure 3.
EXTERNAL VALIDATION
In the external validation, the software correctly segmented and characterised 518 out of 598 regions agreed by consensus, corresponding to an overall diagnostic accuracy of 86.6% (95% CI: 83.7-89.1%). The software performed the best in fibrous plaque (accuracy 97.6% [95% CI: 93.4-99.3%]), followed by lipidic plaque (90.5% [95% CI: 85.2-94.1%]) and calcifications (88.5% [95% CI: 82.4-92.7%]). Cholesterol crystals were also well characterised (accuracy 94.7% [95% CI: 73.5-100%]), but the performance for macrophages was suboptimal (48.1% [95% CI: 37.3-59.0%]) (Figure 2B). The overall diagnostic accuracy was numerically higher in unanimous regions where all three core labs agreed than in those regions where only two core labs agreed: 89.7% (95% CI: 86.7-92.2%) versus 72.7% (95% CI: 63.7-80.2%), p<0.001. When the threshold of overlapping area to define a correct characterisation between the software prediction and the consensus increased from 80% to 90%, diagnostic accuracy decreased slightly but remained high, with 96.4% (95% CI: 92.2-98.5%) for fibrous plaque, 87.2% (95% CI: 81.4-91.3%) for lipidic plaque, and 85.9% (95% CI: 79.5-90.6%) for calcifications.
The diagnostic accuracy was similar in both OCT systems for basic plaque components, 91.9% (95% CI: 88.9-94.2%) in Abbott versus 94.2% (95% CI: 85.6-98.2%) in Terumo (p=0.63), and for all tissue regions 87.5% (95% CI: 84.3-90.1%) in Abbott versus 81.8% (95% CI: 72.4-88.6%) in Terumo (p=0.17).
MODEL RATIONALE AND ANALYSIS TIME
Systematic ablation experiments using the testing data set verified the rationale in the design of the deep convolutional model, including pseudo-3D input, vertical feature bridges, and hybrid loss (Supplementary Table 1).
The median time required for the CNN model to analyse an image pullback with 271 (263-375) cross-sections of 704×704 pixel size was 21.4 (18.6-25.0) seconds, corresponding to an average speed of 0.07±0.01 seconds per cross-section using a laptop equipped with AMD Ryzen 7 and Geforce RTX 2060 graphic card.
Discussion
The following points summarise the key findings of the present study. 1) An AI model based on a deep convolutional neural network for automatic IVOCT plaque characterisation was developed and validated, proving fast computational speed and excellent performance in a real-world series of images. 2) Consensus on coronary tissue characterisation using OCT could be achieved in most of the plaque regions among the three core labs, 81% with unanimity among three core labs and 18% with agreement between two core labs. 3) The AI model performed the best in fibrous plaque, followed by lipidic plaque and calcifications, with diagnostic accuracy of 97.6%, 90.5% and 88.5%, respectively. 4) The plaque burden automatically assessed by the AI model correlated well with the core lab analysis (R2=0.98, p<0.001).
Several studies have previously attempted the automatic characterisation of atherosclerotic plaque on IVOCT using AI4,5. However, these studies developed their models on relatively small data sets with limited diversity, thus entailing problems of generalisability for clinical use. Furthermore, most of these models were validated pursuant to internal analysis within the team, lacking an independent external assessment based on expert consensus. Additionally, the a priori knowledge of spatial continuity along adjacent frames had not been fully exploited and only a few tissue components had been qualitatively characterised hitherto, with limited quantification. In addition to intravascular imaging, the applications of AI in the field of plaque characterisation using cardiac computed tomography (CT) are also expanding. Multiple approaches including machine learning and CNN models have been proposed for automatic calcium detection and scoring using CT images12,13.
From a technical point of view, this novel AI model is unique in many aspects, including the integration of spatial information from contiguous cross-sections to enhance the diagnostic accuracy by means of pseudo-3D input, the incorporation of multi-scale feature forward bridges in both horizontal and vertical directions for better fusion of features, and the use of a hybrid loss function to address challenges in segmentation of inflammatory markers. The rationale for the design of this model was verified on the testing data set (Supplementary Table 1). It is important to note that the average analysis speed for the model is 0.07±0.01 seconds per cross-section while it took the analysts several minutes to annotate one frame in the core lab. It would be more time-demanding for images with complex plaque compositions or suboptimal quality since the specialists need to evaluate cross-sections from several adjacent imaging slices while the model rapidly integrated the information of spatial continuity across frames.
The development of the AI model was focused on clinical applications. In this regard it is important to highlight that the model was trained and validated with a large volume of IVOCT images, encompassing a range of image quality, plaque composition and lesion complexity in a representative population with ischaemic heart disease. This characteristic of the training data set is crucial to guarantee the generalisability of the CNN model. Furthermore, the external validity of the study was retested against a high-quality reference standard, the consensus of three leading international core labs, with excellent diagnostic concordance achieved for most plaque types. Although the data set for external validation was modest in size, it was of different complexity and sufficient to assess the external validity of our findings.
The AI model performed very well in the segmentation of basic plaque components in both internal evaluation and external validation, while the diagnostic accuracy was only modest for markers of high risk and complex plaques. In both categories, the agreement with the expert consensus was better in structures with low versus high attenuation, i.e., the model was more accurate in fibrous than in lipidic tissue, and more accurate in cholesterol crystals than in macrophages. These findings will require a specific appraisal in future studies but might be partially explained by the agreement between the experts in the different categories. The experts tended to disagree the most in structures with high attenuation (lipidic pools, macrophages) or in regions lying very abluminally, where the signal was poor, the quality of the image was lower and therefore the subjective interpretation of the analyst played a greater role. Indeed, the AI model performed significantly better in regions with unanimity than in regions without unanimity (89.7% vs 72.7%, p<0.001).
Of note, the accuracy of the AI model in identifying lipidic plaques (90.5%) was slightly higher than in calcified plaques (88.5%) in the external validation. This was because some lipidic plaques with poor signal penetration, precluding adequate visualisation of boundaries, were flagged with an arch-shaped annotation, which reduced the difficulty for the AI model to meet the standards compared to the calcium plaques delineated with complete boundaries. The accuracy in identifying cholesterol crystals was excellent in the external validation while segmentation performance was modest in the internal testing data set. This might be explained by the small area occupied by cholesterol crystals; a small change in the segmentation border would impact significantly on the Dice index, while preserving good agreement in tissue characterisation. Nevertheless, the model might require further fine-tuning to improve its performance in categories such as macrophages.
To the best of our knowledge, this is the first IVOCT study to report the reproducibility of outlining the IEL. This was a challenging task for IVOCT, because the media often lies beyond IVOCT penetration in thick plaques or is hidden behind lipid-rich pools, causing intense attenuation of the signal1. Nevertheless, recent studies have proven that IVOCT analysts can identify the external elastic lamina (EEL) at >180º of the circumference in 95% of the cross-sections in a core lab setting14. Thus, considering the circular geometry of the arterial structures in the cross-section and the information from continuous frames, the invisible part of IEL and EEL can be reliably extrapolated. As shown in Figure 6, the predicted IEL by our proposed model shows good concordance with an adjacent reference cross-section, even in lesions with a large lipidic pool. This is a key step forward for the quantitative assessment of atherosclerosis by means of IVOCT, with potential prognostic implications, as plaque burden is an independent predictor of future events in non-culprit coronary lesions15.
Figure 6. Predicted IEL by AI model shows good concordance with reference cross-section in plaques with small (A), medium (B, C) and large lipidic pool (D). IEL: internal elastic lamina
Plaque composition also has prognostic value, as large lipidic burden and thin-cap fibroatheroma (TCFA) are associated with higher incidence of periprocedural myocardial infarction16. The current AI model provides comprehensive in vivo analysis of atherosclerotic plaque composition and morphological features of plaque progression or instability in a fully automated fashion, thus providing interventional cardiologists, irrespective of their imaging expertise, with the same level of proficiency as top imaging experts, while sparing time and weariness. The CLIMA trial has recently shown that the identification of macrophages, together with other vulnerable plaque features on OCT, has prognostic value in predicting the population at high risk of acute events17. The AI model is able to identify inflammatory markers automatically along the entire IVOCT pullback rather than in a small region of interest, thus providing a more comprehensive assessment that might potentially result in better risk stratification than conventional image interpretation.
Limitations
This was a study to validate an AI model of automatic segmentation and characterisation of plaque composition, based on the consensus of IVOCT experts. It does not intend to provide a histological validation of IVOCT for plaque characterisation, which has already been achieved18. Of note, histological validation in humans can only be performed by autopsy in cadavers, in patients who died from acute cardiac events. Obtaining histological data from stable coronary lesions in patients dying from non-cardiac causes has proven problematic.
Considering the potential clinical and research applications of the model, the study focused on stable atherosclerotic plaques, excluding lesions with signs of instability (thrombus, plaque rupture, dissection or haematoma). In addition, the presence of metallic stents generates dark trailing shadows on OCT images, jeopardising the interpretation of plaque composition. Thus, cross-sectional images at the stented segment were excluded from the present analysis. The results of the model in unstable plaques and in stented vessel segments should be interpreted with caution and would require further investigation in specific studies.
Conclusions
A novel AI framework for automatic plaque characterisation in IVOCT was developed, providing excellent diagnostic accuracy in both internal and external validation. This model might reduce subjectivity in image interpretation and facilitate IVOCT quantification of plaque composition, with potential applications in research and IVOCT-guided PCI.
Impact on daily practiceAn AI-based model was developed and validated for automatic plaque characterisation on IVOCT. This substantially improved the objectivity and reproducibility of IVOCT quantification. The AI model enables comprehensive plaque characterisation and identification of inflammatory markers, creating an interesting perspective for future studies on plaque progression and risk stratification. The model has the potential to assist IVOCT-guided PCI by tailoring the intervention according to plaque composition and by using internal elastic lamina as reference for stent sizing. |
Funding
This work was supported by the National Key Research and Development Program of China, Natural Science Foundation of China (Grant Number 81871460, 82020108015, 81827806 and 81671763), and by a Science Foundation Ireland Research Professorship Award (RSF 1413).
Conflict of interest statement
Z. Ali reports institutional grants from NIH/NHLBI, Abbott Vascular, and Cardiovascular Systems Inc., personal fees from Amgen, AstraZeneca, and Boston Scientific, and personal equity in Shockwave Medical, outside the submitted work. G. Mintz has received honoraria from Boston Scientific, Philips/Volcano, Medtronic, and Terumo. W. Wijns reports an institutional research grant and honoraria from MicroPort; he is a co-founder of Argonauts, an innovation facilitator. N. Holm has received institutional research grants from Boston Scientific, Biosensors, Abbott, Reva Medical and Medis medical imaging, and speaker fees from Abbott, Terumo, Medis medical imaging and Reva Medical. S. Tu has received research support from Pulse Medical Imaging Technology. The other authors have no conflicts of interest to declare.